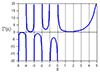
Farrow 2-D Image Interpolation
This snippets applies the Farrow structure to image interpolation, and has been designed
for image co-registration in Synthetic aperture radar (SAR) interferometry. In
co-registration, two images have been sampled in different 2-D grids, and one of them must
be re-sampled in the grid of the other, to allow their joint processing. Usually, this
re-sampling is done using a simple lookup table of the interpolation coefficients.
However this approach takes up a large memory and is inaccurate. The following snippet
performs this interpolation by adapting the Farrow structure to this 2-D setting. There
are, however, some differences with the usual 1-D implementations,
1) The convolution between the 2-D signal and the coefficients is performed using the FFT
convolution. Actually the FFTs are interlaced in both dimensions to get a fast
implementation. (See FarrowInterp2D function.) This is the key of the algorithm.
2) The Farrow coefficients are computed using Chebyshev interpolation and Knab's
pulse. This permits one to update the bandwidths or the number of coefficients in a
simple way. (ChebApproxPolys function.)
The algorithm for this snippet is explained in
[1] J. Selva, J. M. Lopez-Sanchez, 'Efficient Interpolation of SAR Images for
Coregistration in SAR Interferometry,' IEEE Geoscience and Remote Sensing Letters, vol. 4,
n. 3, pp. 411-415, 2007, DOI: http://dx.doi.org/10.1109/LGRS.2007.895961
[2] J. Selva, J. M. Lopez-Sanchez, 'Image coregistration in SAR interferometry only by
means of arithmetic operations', IEEE Geoscience and Remote Sensing Symposium,
2007. IGARSS 2007, DOI: http://dx.doi.org/10.1109/IGARSS.2007.4423854
The Demo function computes a test image formed by a random sum of 2-D sinc pulses and
evaluates the interpolation error. This error can be reduced exponentially by increasing
the truncation index P. However, this increases the image frame in which the interpolation
cannot be performed.
Simply place the following code in a file called "Demo.m".
function Demo
%Author: J. Selva.
%Date: June 2011.
close all
format short g
format compact
T = 1; %Sampling period in both dimensions.
B = 1/(1.223*T); %This is the two-sided bandwidth in both dimensions. It is a typical ...
%value in SAR image co-registration.
P = 18; %Kernel semi-length.
Q = 10; %Polynomial order in Farrow structure.
%The next call to "ChebApproxPolys" computes all Farrow coefficients.
t0 = -P*T-T/2; %The parameters t0, Nt, 2*P+1 specify the 2*P+1 intervals in which Knab's ...
%pulse is to be approximated by polynomials.
Nt = 2*P+1;
Dt = T;
IOut = [-T/2,T/2];
%Perform Chebyshev interpolation. The parameters T, B and P are passed to KnabAPPulse.
K = ChebApproxPolys('KnabAPPulse',t0,Nt,Dt,IOut,40,Q,T,B,P).';
%Evaluate the test image in a regular grid with spacing 1.
Lx = 100;
Ly = 100;
x0 = (0:Lx-1)';
x0 = x0(:,ones(1,Lx));
y0 = x0.';
x0 = x0(:);
y0 = y0(:);
disp('Computing image in a regular grid...')
tic;
Image = reshape(TestImage5(x0,y0,B),[Lx,Ly]);
disp(['(Timing: ',num2str(toc), ' sec.)'])
disp(' ')
clear x0 y0
%Specify NP points on the image at random.
NP = 10000;
x = P*T+rand(1,NP)*(Lx-2*P)*T;
y = P*T+rand(1,NP)*(Ly-2*P)*T;
%Evaluate image directly.
disp('Computing image at selected positions exactly...')
tic;
vRef = TestImage5(x,y,B);
disp(['(Timing: ',num2str(toc), ' sec.)'])
disp(' ')
%Interpolate the image using the 2D-Farrow structure.
disp('Computing image at selected positions using Farrow 2-D...')
tic;
v = Farrow2DInterp(Image,K,x,y);
disp(['(Timing: ',num2str(toc), ' sec.)'])
disp(' ')
%Show the error.
disp(['Maximum interpolation error: ', ...
num2str(20*log10(max(abs(vRef-v))/max(abs(vRef)))),' dB'])
disp(' ')
disp('Display image and interpolation error:')
disp(' ')
%Evaluate the test image in a regular grid with spacing 1.
Lx = 100;
Ly = Lx;
x1 = (P*T:0.5*T:(Lx-P)*T)';
x1 = x1(:,ones(1,length(x1)));
y1 = x1.';
x1 = x1(:);
y1 = y1(:);
tic;
disp('Computing image in over-sampled grid ...')
vRef = TestImage5(x1,y1,B);
disp(['(Timing: ',num2str(toc), ' sec.)'])
disp(' ')
tic;
disp('Computing signal in over-sampled grid using 2-D Farrow ...')
v = Farrow2DInterp(Image,K,x1,y1);
disp(['(Timing: ',num2str(toc), ' sec.)'])
disp(' ')
x1 = (P*T:0.5*T:(Lx-P)*T)';
y1 = x1.';
disp(['Maximum interpolation error: ', ...
num2str(20*log10(max(abs(vRef-v))/max(abs(vRef)))),' dB'])
vRef = reshape(vRef,[length(x1),length(y1)]);
v = reshape(v,[length(x1),length(y1)]);
mesh(x1,y1,abs(vRef))
xlabel('x coordinate')
ylabel('y coordinate')
zlabel('Image''s abs. value')
title('Test Image (abs. value)')
figure
mesh(x1,y1,20*log10(abs(v-vRef)/max(abs(vRef(:)))))
xlabel('x coordinate')
ylabel('y coordinate')
zlabel('Interp. error (dB)')
title('Interpolation error (dB)')
function P = ChebApproxPolys(Func,t0,Nt,Dt,IOut,NPoints,Q,varargin)
%This function constructs Chebyshev interpolation polynomials in a set of consecutive ...
%intervals.
%
% Func is the function to be interpolated.
% t0 is the first instant of the first interval.
% Nt is the number of intervals.
% IOut is the interval in which the output polynomials will be evaluated.
% Dt is the interval width. The intervals are [t0,t0+Dt], [t0+Dt,t0+2*Dt],....
% [t0+(Nt-1)*Dt,t0+Nt*Dt].
% NPoints is the number of points in which Func is evaluated, for each interval.
% Q is the output polynomial order.
% varargin contains extra parameters which are passed to varargin.
%Construct matrix that maps function values to polynomial coefficients.
%The algorithm is that in section 9.8 of
%
% W. H. Press et al., Numerical Recipes in C. Cambridge University
% Press, 1997.
M = MapPolyMatrix([-1,1],IOut,Q) * ChebCoefMatrix(Q) * ...
BlockChebInvMatrix(NPoints,Q);
% Get Chebyshev roots.
r = ChebRoots(NPoints);
r = r(:);
P = zeros(Q,Nt);
%Compute polynomial for each interval.
for k = 0:Nt-1,
v1 = feval(Func,MapIaToIb(r,[-1,1],t0+[k,k+1]*Dt),varargin{:});
P(:,k+1) = double(M * v1(:));
end
function M = MapPolyMatrix(IIn,IOut,Q)
%For a polynomial with Q coefficients which is evaluated in interval IIn, this function ...
%gives the matrix that transforms its coefficients, so that it can be evaluated in the ...
%associated points in interval IOut.
ayx = (IIn(1)-IIn(2))/(IOut(1)-IOut(2));
byx = IIn(1)-ayx*IOut(1);
if byx == 0
M = diag(ayx.^(0:Q-1));
else
M = zeros(Q);
for q = 0:Q-1
for r = 0:q
M(r+1,q+1) = nchoosek(q,r) * ayx^r * byx^(q-r);
end
end
end
function M = ChebCoefMatrix(N)
M = zeros(N);
M(1) = 1;
if N > 1
M(2,2) = 1;
end
for n = 3:N,
M(2:n,n) = 2*M(1:n-1,n-1);
M(1:n-2,n) = M(1:n-2,n) - M(1:n-2,n-2);
end
function M = BlockChebInvMatrix(N,Q)
r = ChebRoots(N);
r= r(:);
M = zeros(N,Q);
if Q >=1
M(:,1) = 1;
end
if Q >= 2
M(:,2) = r;
end
for q = 3:Q,
M(:,q) = 2 * r .* M(:,q-1) - M(:,q-2);
end
M = M.'/N;
M(2:end,:) = M(2:end,:) * 2;
function R = ChebRoots(N)
R = cos(pi*((1:N)-1/2)/N);
function y = MapIaToIb(x,Ia,Ib)
%Linear transformation that maps interval Ia onto interval Ib.
y = (Ib(1)+Ib(2))/2 + (x-(Ia(1)+Ia(2))/2) * ((Ib(2)-Ib(1))/(Ia(2)-Ia(1)));
function [g,BoundAP] = KnabAPPulse(t,T,B,P)
%Knab interpolation pulse in
%
% J. J. Knab, 'Interpolation of band-limited functions using the Approximate
% Prolate series', IEEE Transactions on Information Theory, vol. IT-25,
% no. 6, pp. 717-720, Nov 1979.
%
%Grid points -P*T:T:P*T.
%Interpolation interval: [-T/2,T/2].
g = sinc(t/T) .* sinc((1/T-B)*sqrt(t.^2-(P*T)^2)) / sinc(j*(1-B*T)*P);
if nargout == 2
BoundAP = 1/sinh(P*pi*(1-B*T));
end
function v = TestImage5(x,y,B)
%Test image formed by a sum of random sincs.
Seed = rand('state');
rand('state',362436069);
L = 100; %Image size in both dimensions.
Np = 5000;
p = rand(Np,2)*L;
A = exp(j*2*pi*rand(Np,1));
rand('state',Seed);
v = zeros(size(x));
for k = 1:length(x(:))
v(k) = A(:).' * (sinc(B*(x(k)-p(:,1))) .* sinc(B*(y(k)-p(:,2))));
end
function vx = Farrow2DInterp(Image,Kernel,x,y)
%Farrow interpolation in two dimensions using interlaced FFTs.
vxSize = size(x);
[LK,NK] = size(Kernel);
[Lx,Ly] = size(Image);
nx = round(x);
ny = round(y);
ux = x-nx;
uy = y-ny;
np = 1+nx+ny*Lx;
np = np(:);
clear x y nx ny
P = (LK-1)/2;
NFFTx = 2^ceil(log2(Lx+LK-1));
NFFTy = 2^ceil(log2(Ly+LK-1));
KernelFx = fft(Kernel,NFFTx);
KernelFy = fft(Kernel,NFFTy);
ImageFx = fft(Image,NFFTx);
vx = zeros(size(np))+j*zeros(size(np));
for kx = NK:-1:1,
ImageCx = ifft(ImageFx .* KernelFx(:,kx(ones(1,Ly))),NFFTx);
ImageCx = ImageCx(1+P:Lx+LK-1-P,:);
ImageCxFy = fft(ImageCx,NFFTy,2);
vy = zeros(size(np))+j*zeros(size(np));
for ky = NK:-1:1,
ImageCxCy = ifft(ImageCxFy .* KernelFy(:,ky(ones(1,Lx))).',NFFTy,2);
ImageCxCy = ImageCxCy(:,1+P:Ly+LK-1-P);
vy(:) = vy(:) .* uy(:) + ImageCxCy(np);
end
vx(:) = vx(:) .* ux(:) + vy(:);
end
vx = reshape(vx,vxSize);