Steve-
> Thanks for the quick responses. But the windows check
the pixel values every
> frame so I think it is measuring the movement of gaps between successive
> images. The output has two signals; one is a *roughly* delayed version of
> the first signal, showing there is some correlation between the windows.
But
> the xcorr function doesn't seem to be giving me the right delay, because
by
> eye, the graphs indicate that it's ~5 frames on the x axis (time), but
it
> keeps spitting out the total amount of frames as the delay.
At least in my understanding, I don't see the value of any type of "time
axis" readout -- you already know the frame
rate.
If you apply Xcorr() across a series of frames then you would expect to see a
resulting series of frames with a peak
(like a sinx/x feature using simple test images) in each one. I.e. where the
particle (or target image) is matched,
or highly correlated, within each frame. It seems to me that you have to
consider the pixel shift between each
sucessive frame in order to obtain a meaningful velocity result. Maybe if you
look at Xcorr() output and find the
relative changes between x and y positions of each peak, that's useful.
-Jeff
> On Mon, Aug 17, 2009 at 4:14 PM, Jeff Brower
wrote:
>
>> Steve-
>>
>> Sorry, that should say ... you're not actually measuring the movement of
a
>> particle between *successive* images.
>>
>> -Jeff
>>
>> ------------------ Original Message ------------------
>> Subject: Re: [matlab] Time Delay Measurement in MATLAB
>> From: "Jeff Brower"
>> Date: Mon, August 17, 2009 6:07 pm
>> To: "Steve"
>> Cc: m...
>> -------------------
>>
>> Steve-
>>
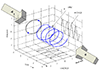
>> > I am only cross correlating two windows of the images, mainly the edges.
>> The windows sense the pixel changes so it
>> can track lighter particles (due to gaps between red blood cells) moving
>> through both windows, and the cross
>> correlation should measure the time it takes to go from one window to the
>> next.
>>
>> "...the cross correlation should measure the time it takes to go from one
>> window to the next."
>>
>> At first glance, this sounds like it explains why you always get the
number
>> of frames as a delay value. You're not
>> actually measuring the movement of a particle within one image.
>>
>> -Jeff
>>
>> > On Mon, Aug 17, 2009 at 8:39 AM, Jeff Brower
>> wrote:
>> >
>> >> Steve-
>> >>
>> >> Are you cross correlating the entire image, or only the extracted
>> particle (or other marker) from the fluid flow? If
>> >> the entire image, then are there multiple particles? If so should they
>> all be moving at the same speed?
>> >>
>> >> One way to test your code would be to create simulated images; for
>> example, place a pattern (circle, etc) in the
>> >> center of image 1, then move it slightly over and down in image 2, then
>> run xcorr(). You should get an x-y offset in
>> >> pixels.
>> >>
>> >> -Jeff
>> >>
>> >> > I am trying to measure fluid velocity in an AVI file, and am doing
>> this by converting each frame to an image and
>> measuring the mean intensity of the pixels in two different windows spaced
>> out. Now that I have two different
>> signals
>> >> > that should be theoretically time delayed, I need to cross
>> >> > correlate them to see the time delay, but I am having trouble with
>> xcorr. I seem to obtain the signals alright,
>> but the xcorr always says that the delay is the number of frames I test
>> (when t=1:30, the delay is 30, when
>> t=1:400, the delay is 400). My code is posted
>> >> below:
>> >> >
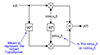
>> >> > %Splits frames of AVI into grayscale images. Then it takes pixel
>> >> intensity
>> >> > %averages of the windows and inputs into two arrays AllIntensity1 and
>> %AllIntensity2. Then it cross correlates
>> the signals.
>> >> >
>> >> > A=mmreader('new.avi') %Reads Avi movie, places into A.mov
for
>> Matlab's
>> >> %viewing pleasure.
>> >> >
>> >> > for t=1:30
>> >> > im = read( A, t);%reads sequential frames
>> >> > imgray=rgb2gray(im);%converts read images into grayscale
>> >> >
>> >> > %Chooses windows in first iteration only
>> >> > if (t==1)
>> >> > BW1=roipoly(imgray);
>> >> > BW2=roipoly(imgray);
>> >> > end
>> >> > %computes mean intensity for each window separately, placing in
>> two
>> >> %arrays
>> >> > Stats=regionprops(BW1,imgray,'MeanIntensity');
>> >> > AllIntensity1(t)=Stats.MeanIntensity;
>> >> > Stats=regionprops(BW2,imgray,'MeanIntensity');
>> >> > AllIntensity2(t)=Stats.MeanIntensity;
>> >> >
>> >> > end
>> >> > AllIntensity2=[100 100 103 105 103 100 AllIntensity1(1:24)];
>> >> >
>> >> > %%%%%%%%%%%%%%%%%%%%%%%%%%%Cross Correlation
>> >> Begins%%%%%%%%%%%%%%%%%%%%%%%%
>> >> xc=xcorr(AllIntensity1,AllIntensity2,'coeff');
>> >> > [a,delay]=max(xc);
>> >> > a
>> >> > delay
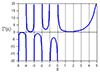
>> >> > subplot(311),plot(AllIntensity1)
>> >> > subplot(312),plot(AllIntensity2)
>> >> > subplot(313),plot(xc(1:59))
>> >> >
>> >> > Any Ideas?? Thanks!
>> >> > Steve
>
Re: [Fwd: Re: Time Delay Measurement in MATLAB]
Started by ●August 17, 2009