I'm attempting to demonstrate some symmetries with real 1d and 2d
signals,
but finding that what I expect to happen (see Proakis' DSP book section
7.2,
or gonzalez & woods page 242) is not what I get.
I've written up a simple example demonstrating the problems I'm
seeing.
For reference, here are the properties I'm attempting to demonstrate:
Given a real-valued sequence f(x), and its DFT F(u) (where * denotes the
complex conjugate):
:- F(u) = F*(-u) => F*(u) = F(-u)
:- f(-x) <=> F(-u) = F*(u)
The code below uses x and X, defined as follows:
-=-=-=-=-=-=-=-=-=-=-=-=-
x = randn(30,1);
X = fft(x,32);
-=-=-=-=-=-=-=-=-=-=-=-=-
Here's the code (comments explain what is expected and some nuances):
% Testing frequency reversal. Similar results occur if fftshift is used,
% though I expected as much since I expect sequence reversal to be
% shift invariant.
>> [[x;0;0] ifft(flipdim(X,1))]
ans 0.3899 0.3899
0.0880 0.0000 - 0.0000i
-0.6355 -0.0000 + 0.0000i
-0.5596 0.7928 - 0.5297i
0.4437 -0.1846 + 0.1846i
-0.9499 -0.7496 + 1.1219i
0.7812 -0.4318 + 1.0425i
0.5690 -0.0080 + 0.0404i
-0.8217 0 + 1.2173i
-0.2656 -0.0109 - 0.0547i
-1.1878 -0.5635 - 1.3604i
-2.2023 0.5866 + 0.8780i
0.9863 0.8385 + 0.8385i
-0.5186 0.3113 + 0.2080i
0.3274 0.8750 + 0.3625i
0.2341 0.9847 + 0.1959i
0.0215 -0.0215
-1.0039 -0.2296 + 0.0457i
-0.9471 -0.3024 + 0.1253i
-0.3744 0.4312 - 0.2881i
-1.1859 -0.6974 + 0.6974i
-1.0559 1.2235 - 1.8312i
1.4725 0.4545 - 1.0974i
0.0557 0.0518 - 0.2605i
-1.2173 0 - 0.8217i
-0.0412 0.1110 + 0.5580i
-1.1283 0.2989 + 0.7217i
-1.3493 -0.5277 - 0.7898i
-0.2611 0.3137 + 0.3137i
0.9535 -0.4653 - 0.3109i
0 -0.5871 - 0.2432i
0 0.0863 + 0.0172i
% Testing conjugation in the frequency domain
% The sequence /is/ reversed, but is shifted circularly. Why?
% Exact same thing occurs if fftshift/ifftshift are used, though this should
be expected
>> [[x;0;0] ifft(conj(X))]
ans 0.1286 0.1286
0.6565 0.0000
-1.1678 0
-0.4606 -1.2013
-0.2624 -0.8542
-1.2132 0.6630
-1.3194 -0.2463
0.9312 -0.8468
0.0112 -0.5465
-0.6451 1.4885
0.8057 0.4978
0.2316 -0.8436
-0.9898 -0.3306
1.3396 -0.0729
0.2895 -1.2919
1.4789 -0.6841
1.1380 1.1380
-0.6841 1.4789
-1.2919 0.2895
-0.0729 1.3396
-0.3306 -0.9898
-0.8436 0.2316
0.4978 0.8057
1.4885 -0.6451
-0.5465 0.0112
-0.8468 0.9312
-0.2463 -1.3194
0.6630 -1.2132
-0.8542 -0.2624
-1.2013 -0.4606
0 -1.1678
0 0.6565
% Obviously conj(flipdim(...)) will be corrupted as well, but here it is
anyway:
>> [[x;0;0] ifft(conj(flipdim(X,1)))]
ans 0.1286 0.1286
0.6565 0.6439 - 0.1281i
-1.1678 -1.0789 + 0.4469i
-0.4606 -0.3830 + 0.2559i
-0.2624 -0.1856 + 0.1856i
-1.2132 -0.6740 + 1.0087i
-1.3194 -0.5049 + 1.2190i
0.9312 0.1817 - 0.9133i
0.0112 0 - 0.0112i
-0.6451 0.1259 + 0.6327i
0.8057 -0.3083 - 0.7444i
0.2316 -0.1287 - 0.1926i
-0.9898 0.6999 + 0.6999i
1.3396 -1.1138 - 0.7442i
0.2895 -0.2675 - 0.1108i
1.4789 -1.4505 - 0.2885i
1.1380 -1.1380
-0.6841 0.6710 - 0.1335i
-1.2919 1.1936 - 0.4944i
-0.0729 0.0606 - 0.0405i
-0.3306 0.2338 - 0.2338i
-0.8436 0.4687 - 0.7015i
0.4978 -0.1905 + 0.4599i
1.4885 -0.2904 + 1.4599i
-0.5465 0 - 0.5465i
-0.8468 -0.1652 - 0.8305i
-0.2463 -0.0943 - 0.2276i
0.6630 0.3684 + 0.5513i
-0.8542 -0.6040 - 0.6040i
-1.2013 -0.9989 - 0.6674i
0 0.0000 - 0.0000i
0 0.0000
The big question is: why is this occurring? I've performed similar
tests
in R and I see similar results. I've also tried this with different
types
of sequences (random, sequential integers, constant sequences, etc) with
similar results. The real part of the ifft starts out very near the
original sequence but between 0 and N/2 the values deviate fairly fast,
converge again around N/2 then diverge between N/2 and N.
Also, I've found the real part is far more accurate if I use a much
larger
DFT (2048, 4096, etc) but at the obvious expense of performing a very
expensive DFT.
Any comments/suggestions?
-Brian
issues testing DFT symmetries
Started by ●January 20, 2010
Reply by ●January 20, 20102010-01-20
Brian-
> I'm attempting to demonstrate some symmetries with real 1d and 2d signals,
> but finding that what I expect to happen (see Proakis' DSP book section 7.2,
> or gonzalez & woods page 242) is not what I get.
>
> I've written up a simple example demonstrating the problems I'm seeing.
> For reference, here are the properties I'm attempting to demonstrate:
>
> Given a real-valued sequence f(x), and its DFT F(u) (where * denotes the
> complex conjugate):
> :- F(u) = F*(-u) => F*(u) = F(-u)
> :- f(-x) <=> F(-u) = F*(u)
>
> The code below uses x and X, defined as follows:
>
> -=-=-=-=-=-=-=-=-=-=-=-=-
> x = randn(30,1);
> X = fft(x,32);
> -=-=-=-=-=-=-=-=-=-=-=-=-
>
> Here's the code (comments explain what is expected and some nuances):
>
> % Testing frequency reversal. Similar results occur if fftshift is used,
> % though I expected as much since I expect sequence reversal to be
> % shift invariant.
>>> [[x;0;0] ifft(flipdim(X,1))]
I may be asking too quick, but if you do X = fft(x) and do anything to create X' from X so they're even slightly
different, then you do y = ifft(X'), then the result is x <> y (because Fourier transform is unique). Why can't you
just compare X and X'? For example, X - flipdim(X,1): does that give exactly zero?
Another quick thought is that ifft(X) (or ifft(X flipped)) is going to give you 32 points, not 30. Isn't that an
issue? You did mention DFT, so why not control the FFT size to be the exact length of your input sequence?
-Jeff
> ans > 0.3899 0.3899
> 0.0880 0.0000 - 0.0000i
> -0.6355 -0.0000 + 0.0000i
> -0.5596 0.7928 - 0.5297i
> 0.4437 -0.1846 + 0.1846i
> -0.9499 -0.7496 + 1.1219i
> 0.7812 -0.4318 + 1.0425i
> 0.5690 -0.0080 + 0.0404i
> -0.8217 0 + 1.2173i
> -0.2656 -0.0109 - 0.0547i
> -1.1878 -0.5635 - 1.3604i
> -2.2023 0.5866 + 0.8780i
> 0.9863 0.8385 + 0.8385i
> -0.5186 0.3113 + 0.2080i
> 0.3274 0.8750 + 0.3625i
> 0.2341 0.9847 + 0.1959i
> 0.0215 -0.0215
> -1.0039 -0.2296 + 0.0457i
> -0.9471 -0.3024 + 0.1253i
> -0.3744 0.4312 - 0.2881i
> -1.1859 -0.6974 + 0.6974i
> -1.0559 1.2235 - 1.8312i
> 1.4725 0.4545 - 1.0974i
> 0.0557 0.0518 - 0.2605i
> -1.2173 0 - 0.8217i
> -0.0412 0.1110 + 0.5580i
> -1.1283 0.2989 + 0.7217i
> -1.3493 -0.5277 - 0.7898i
> -0.2611 0.3137 + 0.3137i
> 0.9535 -0.4653 - 0.3109i
> 0 -0.5871 - 0.2432i
> 0 0.0863 + 0.0172i
>
> % Testing conjugation in the frequency domain
> % The sequence /is/ reversed, but is shifted circularly. Why?
> % Exact same thing occurs if fftshift/ifftshift are used, though this should
> be expected
>>> [[x;0;0] ifft(conj(X))]
> ans > 0.1286 0.1286
> 0.6565 0.0000
> -1.1678 0
> -0.4606 -1.2013
> -0.2624 -0.8542
> -1.2132 0.6630
> -1.3194 -0.2463
> 0.9312 -0.8468
> 0.0112 -0.5465
> -0.6451 1.4885
> 0.8057 0.4978
> 0.2316 -0.8436
> -0.9898 -0.3306
> 1.3396 -0.0729
> 0.2895 -1.2919
> 1.4789 -0.6841
> 1.1380 1.1380
> -0.6841 1.4789
> -1.2919 0.2895
> -0.0729 1.3396
> -0.3306 -0.9898
> -0.8436 0.2316
> 0.4978 0.8057
> 1.4885 -0.6451
> -0.5465 0.0112
> -0.8468 0.9312
> -0.2463 -1.3194
> 0.6630 -1.2132
> -0.8542 -0.2624
> -1.2013 -0.4606
> 0 -1.1678
> 0 0.6565
>
> % Obviously conj(flipdim(...)) will be corrupted as well, but here it is
> anyway:
>>> [[x;0;0] ifft(conj(flipdim(X,1)))]
> ans > 0.1286 0.1286
> 0.6565 0.6439 - 0.1281i
> -1.1678 -1.0789 + 0.4469i
> -0.4606 -0.3830 + 0.2559i
> -0.2624 -0.1856 + 0.1856i
> -1.2132 -0.6740 + 1.0087i
> -1.3194 -0.5049 + 1.2190i
> 0.9312 0.1817 - 0.9133i
> 0.0112 0 - 0.0112i
> -0.6451 0.1259 + 0.6327i
> 0.8057 -0.3083 - 0.7444i
> 0.2316 -0.1287 - 0.1926i
> -0.9898 0.6999 + 0.6999i
> 1.3396 -1.1138 - 0.7442i
> 0.2895 -0.2675 - 0.1108i
> 1.4789 -1.4505 - 0.2885i
> 1.1380 -1.1380
> -0.6841 0.6710 - 0.1335i
> -1.2919 1.1936 - 0.4944i
> -0.0729 0.0606 - 0.0405i
> -0.3306 0.2338 - 0.2338i
> -0.8436 0.4687 - 0.7015i
> 0.4978 -0.1905 + 0.4599i
> 1.4885 -0.2904 + 1.4599i
> -0.5465 0 - 0.5465i
> -0.8468 -0.1652 - 0.8305i
> -0.2463 -0.0943 - 0.2276i
> 0.6630 0.3684 + 0.5513i
> -0.8542 -0.6040 - 0.6040i
> -1.2013 -0.9989 - 0.6674i
> 0 0.0000 - 0.0000i
> 0 0.0000
>
> The big question is: why is this occurring? I've performed similar tests
> in R and I see similar results. I've also tried this with different types
> of sequences (random, sequential integers, constant sequences, etc) with
> similar results. The real part of the ifft starts out very near the
> original sequence but between 0 and N/2 the values deviate fairly fast,
> converge again around N/2 then diverge between N/2 and N.
>
> Also, I've found the real part is far more accurate if I use a much larger
> DFT (2048, 4096, etc) but at the obvious expense of performing a very
> expensive DFT.
>
> Any comments/suggestions?
>
> -Brian
>
> I'm attempting to demonstrate some symmetries with real 1d and 2d signals,
> but finding that what I expect to happen (see Proakis' DSP book section 7.2,
> or gonzalez & woods page 242) is not what I get.
>
> I've written up a simple example demonstrating the problems I'm seeing.
> For reference, here are the properties I'm attempting to demonstrate:
>
> Given a real-valued sequence f(x), and its DFT F(u) (where * denotes the
> complex conjugate):
> :- F(u) = F*(-u) => F*(u) = F(-u)
> :- f(-x) <=> F(-u) = F*(u)
>
> The code below uses x and X, defined as follows:
>
> -=-=-=-=-=-=-=-=-=-=-=-=-
> x = randn(30,1);
> X = fft(x,32);
> -=-=-=-=-=-=-=-=-=-=-=-=-
>
> Here's the code (comments explain what is expected and some nuances):
>
> % Testing frequency reversal. Similar results occur if fftshift is used,
> % though I expected as much since I expect sequence reversal to be
> % shift invariant.
>>> [[x;0;0] ifft(flipdim(X,1))]
I may be asking too quick, but if you do X = fft(x) and do anything to create X' from X so they're even slightly
different, then you do y = ifft(X'), then the result is x <> y (because Fourier transform is unique). Why can't you
just compare X and X'? For example, X - flipdim(X,1): does that give exactly zero?
Another quick thought is that ifft(X) (or ifft(X flipped)) is going to give you 32 points, not 30. Isn't that an
issue? You did mention DFT, so why not control the FFT size to be the exact length of your input sequence?
-Jeff
> ans > 0.3899 0.3899
> 0.0880 0.0000 - 0.0000i
> -0.6355 -0.0000 + 0.0000i
> -0.5596 0.7928 - 0.5297i
> 0.4437 -0.1846 + 0.1846i
> -0.9499 -0.7496 + 1.1219i
> 0.7812 -0.4318 + 1.0425i
> 0.5690 -0.0080 + 0.0404i
> -0.8217 0 + 1.2173i
> -0.2656 -0.0109 - 0.0547i
> -1.1878 -0.5635 - 1.3604i
> -2.2023 0.5866 + 0.8780i
> 0.9863 0.8385 + 0.8385i
> -0.5186 0.3113 + 0.2080i
> 0.3274 0.8750 + 0.3625i
> 0.2341 0.9847 + 0.1959i
> 0.0215 -0.0215
> -1.0039 -0.2296 + 0.0457i
> -0.9471 -0.3024 + 0.1253i
> -0.3744 0.4312 - 0.2881i
> -1.1859 -0.6974 + 0.6974i
> -1.0559 1.2235 - 1.8312i
> 1.4725 0.4545 - 1.0974i
> 0.0557 0.0518 - 0.2605i
> -1.2173 0 - 0.8217i
> -0.0412 0.1110 + 0.5580i
> -1.1283 0.2989 + 0.7217i
> -1.3493 -0.5277 - 0.7898i
> -0.2611 0.3137 + 0.3137i
> 0.9535 -0.4653 - 0.3109i
> 0 -0.5871 - 0.2432i
> 0 0.0863 + 0.0172i
>
> % Testing conjugation in the frequency domain
> % The sequence /is/ reversed, but is shifted circularly. Why?
> % Exact same thing occurs if fftshift/ifftshift are used, though this should
> be expected
>>> [[x;0;0] ifft(conj(X))]
> ans > 0.1286 0.1286
> 0.6565 0.0000
> -1.1678 0
> -0.4606 -1.2013
> -0.2624 -0.8542
> -1.2132 0.6630
> -1.3194 -0.2463
> 0.9312 -0.8468
> 0.0112 -0.5465
> -0.6451 1.4885
> 0.8057 0.4978
> 0.2316 -0.8436
> -0.9898 -0.3306
> 1.3396 -0.0729
> 0.2895 -1.2919
> 1.4789 -0.6841
> 1.1380 1.1380
> -0.6841 1.4789
> -1.2919 0.2895
> -0.0729 1.3396
> -0.3306 -0.9898
> -0.8436 0.2316
> 0.4978 0.8057
> 1.4885 -0.6451
> -0.5465 0.0112
> -0.8468 0.9312
> -0.2463 -1.3194
> 0.6630 -1.2132
> -0.8542 -0.2624
> -1.2013 -0.4606
> 0 -1.1678
> 0 0.6565
>
> % Obviously conj(flipdim(...)) will be corrupted as well, but here it is
> anyway:
>>> [[x;0;0] ifft(conj(flipdim(X,1)))]
> ans > 0.1286 0.1286
> 0.6565 0.6439 - 0.1281i
> -1.1678 -1.0789 + 0.4469i
> -0.4606 -0.3830 + 0.2559i
> -0.2624 -0.1856 + 0.1856i
> -1.2132 -0.6740 + 1.0087i
> -1.3194 -0.5049 + 1.2190i
> 0.9312 0.1817 - 0.9133i
> 0.0112 0 - 0.0112i
> -0.6451 0.1259 + 0.6327i
> 0.8057 -0.3083 - 0.7444i
> 0.2316 -0.1287 - 0.1926i
> -0.9898 0.6999 + 0.6999i
> 1.3396 -1.1138 - 0.7442i
> 0.2895 -0.2675 - 0.1108i
> 1.4789 -1.4505 - 0.2885i
> 1.1380 -1.1380
> -0.6841 0.6710 - 0.1335i
> -1.2919 1.1936 - 0.4944i
> -0.0729 0.0606 - 0.0405i
> -0.3306 0.2338 - 0.2338i
> -0.8436 0.4687 - 0.7015i
> 0.4978 -0.1905 + 0.4599i
> 1.4885 -0.2904 + 1.4599i
> -0.5465 0 - 0.5465i
> -0.8468 -0.1652 - 0.8305i
> -0.2463 -0.0943 - 0.2276i
> 0.6630 0.3684 + 0.5513i
> -0.8542 -0.6040 - 0.6040i
> -1.2013 -0.9989 - 0.6674i
> 0 0.0000 - 0.0000i
> 0 0.0000
>
> The big question is: why is this occurring? I've performed similar tests
> in R and I see similar results. I've also tried this with different types
> of sequences (random, sequential integers, constant sequences, etc) with
> similar results. The real part of the ifft starts out very near the
> original sequence but between 0 and N/2 the values deviate fairly fast,
> converge again around N/2 then diverge between N/2 and N.
>
> Also, I've found the real part is far more accurate if I use a much larger
> DFT (2048, 4096, etc) but at the obvious expense of performing a very
> expensive DFT.
>
> Any comments/suggestions?
>
> -Brian
>
Reply by ●January 20, 20102010-01-20
>>
>> Here's the code (comments explain what is expected and some nuances):
>>
>> % Testing frequency reversal. Similar results occur if fftshift is used,
>> % though I expected as much since I expect sequence reversal to be
>> % shift invariant.
>>>> [[x;0;0] ifft(flipdim(X,1))]
> I may be asking too quick, but if you do X = fft(x) and do anything to create X'
> from X so they're even slightly
> different, then you do y = ifft(X'), then the result is x <>> y (because
> Fourier transform is unique). Why can't you
> just compare X and X'? For example, X - flipdim(X,1): does that give exactly
> zero?
>
> Another quick thought is that ifft(X) (or ifft(X flipped)) is going to give you
> 32 points, not 30. Isn't that an
> issue? You did mention DFT, so why not control the FFT size to be the exact
> length of your input sequence?
>
> -Jeff
Well, I realized today I made one small mistake in my assumptions. I do see different results when using fftshift. A simple example demonstrates why I was wrong:
Original sequence:
abcd
After fftshift:
cdab
Sequence reversal:
badc
fftshift again:
dcab
Even if I use fftshift, though, the results are still wrong.
One thing I just discovered while tinkering, though, is this:
% Note that I'm computing the magnitude of the inverse,
% as opposed to just an ifft. The ifft is still wrong.
x = (1:30)';
X = fft(x,32);
[[x;0;0] abs(ifft(flipdim(X,1)))]
ans
1.0000 1.0000
2.0000 0.0000
3.0000 0.0000
4.0000 30.0000
5.0000 29.0000
6.0000 28.0000
7.0000 27.0000
8.0000 26.0000
9.0000 25.0000
10.0000 24.0000
11.0000 23.0000
12.0000 22.0000
13.0000 21.0000
14.0000 20.0000
15.0000 19.0000
16.0000 18.0000
17.0000 17.0000
18.0000 16.0000
19.0000 15.0000
20.0000 14.0000
21.0000 13.0000
22.0000 12.0000
23.0000 11.0000
24.0000 10.0000
25.0000 9.0000
26.0000 8.0000
27.0000 7.0000
28.0000 6.0000
29.0000 5.0000
30.0000 4.0000
0.0000 3.0000
0.0000 2.0000
Now I'm baffled. Flipping in the frequency domain with fftshift (and unshifting afterward), taking the inverse transform, then computing the magnitude of the sequence gives [almost] what I expect, but I shouldn't have to take the magnitude of the sequence. Furthermore, it's circularly shifting by 1 still.
-Brian
>> Here's the code (comments explain what is expected and some nuances):
>>
>> % Testing frequency reversal. Similar results occur if fftshift is used,
>> % though I expected as much since I expect sequence reversal to be
>> % shift invariant.
>>>> [[x;0;0] ifft(flipdim(X,1))]
> I may be asking too quick, but if you do X = fft(x) and do anything to create X'
> from X so they're even slightly
> different, then you do y = ifft(X'), then the result is x <>> y (because
> Fourier transform is unique). Why can't you
> just compare X and X'? For example, X - flipdim(X,1): does that give exactly
> zero?
>
> Another quick thought is that ifft(X) (or ifft(X flipped)) is going to give you
> 32 points, not 30. Isn't that an
> issue? You did mention DFT, so why not control the FFT size to be the exact
> length of your input sequence?
>
> -Jeff
Well, I realized today I made one small mistake in my assumptions. I do see different results when using fftshift. A simple example demonstrates why I was wrong:
Original sequence:
abcd
After fftshift:
cdab
Sequence reversal:
badc
fftshift again:
dcab
Even if I use fftshift, though, the results are still wrong.
One thing I just discovered while tinkering, though, is this:
% Note that I'm computing the magnitude of the inverse,
% as opposed to just an ifft. The ifft is still wrong.
x = (1:30)';
X = fft(x,32);
[[x;0;0] abs(ifft(flipdim(X,1)))]
ans
1.0000 1.0000
2.0000 0.0000
3.0000 0.0000
4.0000 30.0000
5.0000 29.0000
6.0000 28.0000
7.0000 27.0000
8.0000 26.0000
9.0000 25.0000
10.0000 24.0000
11.0000 23.0000
12.0000 22.0000
13.0000 21.0000
14.0000 20.0000
15.0000 19.0000
16.0000 18.0000
17.0000 17.0000
18.0000 16.0000
19.0000 15.0000
20.0000 14.0000
21.0000 13.0000
22.0000 12.0000
23.0000 11.0000
24.0000 10.0000
25.0000 9.0000
26.0000 8.0000
27.0000 7.0000
28.0000 6.0000
29.0000 5.0000
30.0000 4.0000
0.0000 3.0000
0.0000 2.0000
Now I'm baffled. Flipping in the frequency domain with fftshift (and unshifting afterward), taking the inverse transform, then computing the magnitude of the sequence gives [almost] what I expect, but I shouldn't have to take the magnitude of the sequence. Furthermore, it's circularly shifting by 1 still.
-Brian
Reply by ●January 21, 20102010-01-21
> Now I'm baffled. Flipping in the frequency
domain with fftshift (and
> unshifting afterward), taking the inverse transform, then computing the
> magnitude of the sequence gives [almost] what I expect, but I shouldn't have to
> take the magnitude of the sequence. Furthermore, it's circularly shifting by 1
> still.
I got a minute to tinker with this some more. It looks as though something I'm doing (incorrectly?) is roughly equivalent to multiplying the reversed (and circularly shifted) sequence by a complex sequence with linear phase.
I ran the following code in R:
x <- c(1:32, 0, 0);
X <- fft(x);
X <- rev(X[c(17:32, 1:16)]);
X <- X[c(17:32, 1:16)];
plot(fft(X, inverse=TRUE));
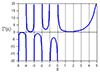
... and this is what the plot looks like:
http://upload.decay.org/720_ULwdo1DU227HT6FP_phase.jpg
If anyone has some insight into this I would love to hear it.
-Brian
> unshifting afterward), taking the inverse transform, then computing the
> magnitude of the sequence gives [almost] what I expect, but I shouldn't have to
> take the magnitude of the sequence. Furthermore, it's circularly shifting by 1
> still.
I got a minute to tinker with this some more. It looks as though something I'm doing (incorrectly?) is roughly equivalent to multiplying the reversed (and circularly shifted) sequence by a complex sequence with linear phase.
I ran the following code in R:
x <- c(1:32, 0, 0);
X <- fft(x);
X <- rev(X[c(17:32, 1:16)]);
X <- X[c(17:32, 1:16)];
plot(fft(X, inverse=TRUE));
... and this is what the plot looks like:
http://upload.decay.org/720_ULwdo1DU227HT6FP_phase.jpg
If anyone has some insight into this I would love to hear it.
-Brian